R and Rstudio
R is a popular statistical programming language for data science and analysis. To use R on Hyak, we rely on Apptainer and Docker containers to deploy R. You might find a refresher on containers and modules helpful before following these instructions.
User Environment
If you use a non-custom R container you'll likely run install.packages() at some point. Usually on a non-shared platform like your local setup (where you have full administrative privileges) R will install things into central paths. On Hyak, R package libraries are usually installed by default in the user's Home directory, which can be problematic due to the 10GB disk storage limit. If this default setting isn't changed, users can quickly run out of storage and inodes in their Home directory and need to re-configure their R environment.
Instead of waiting for the inevitable, we will direct R to install package libraries in a directory we choose where storage isn't limited. This might be your lab groups directory under /gscratch/ or a directory you creaed under you UW Net ID, like, /gscratch/scrubbed/UWNetID. Click here to review storage on Hyak.
Remember to replace the word UWNetID in the paths below with YOUR username/UWNetID.
Specify user library paths by editing or creating a configuration file called .Renviron in your Home directory. Use nano or vim to designate the location of your R package libraries. The contents of the file should be something like the following example.
$ cat ~/.Renviron
R_LIBS="/gscratch/scrubbed/UWNetID/R/"
Remember if the directory you want to use doesn't exist yet, R will send an error message. If you want to create a directory for yourself in /gscratch/scrubbed use the following command:
mkdir /gscratch/scrubbed/UWNetID/
# remember to replace the word `UWNetID` above with YOUR username/UWNetID
And then create a directory to store your R package libraries called R:
mkdir /gscratch/scrubbed/UWNetID/R
# remember to replace the word `UWNetID` above with YOUR username/UWNetID
Now R will install packages in your designated directory instead of your Home directory, and you will avoid disk storage management issues later on. Don't forget to bind your designated directory when you run R, which we cover below and you can learn more about in the container tutorial.
If you plan on using multiple R versions you will want to set R_LIBS appropriately with each different container (i.e., R version) used so packages compiled against one version of R don't conflict with another. Using sub-folders with names matching that version of R is sufficient.
Containers from Rocker
The Rocker Project on Docker hub hosts many containers that were prepared by the developers of R and many include various package collections. The Rocker Project on Docker hub hosts many containers that were prepared by the developers of R
(https://hub.docker.com/u/rocker). In this part of the guide, we will walk you through a few of the options and show you how to set them up for your usage on klone.
R-base Container
Let's say we wanted to use the most up-to-date version of base R from the Rocker Project on Docker hub [More information here.]. There are many other versions are R available on Docker hub, and we encourage you to explore them to find the version that fits the needs of your research project. Explore versions here.
First start an interactive job on a compute node. Building containers is not a login-node approved activity. The following command will request a single CPU on the ckpt parition with 16GB of RAM for 2 hours. If your lab group owns Hyak resources, you might be able to change --partition=ckpt to --partition=compute for priority access to a node. Find out which resources you can use with the hyakalloc command.
salloc --partition=ckpt --cpus-per-task=1 --mem=16G --time=2:00:00
Pull the container from Docker hub with Apptainer.
apptainer pull docker://rocker/r-base
INFO: Converting OCI blobs to SIF format
INFO: Starting build...
Getting image source signatures
Skipping fetch of repeat blob sha256:ecd924c7226f314c16de965258f37da4aa990c07e494be1116af512706138401
Skipping fetch of repeat blob sha256:4fec22ce03e6be2d27efb3e9a90be68b14183859abf0864518e2581aa49fb8f5
Skipping fetch of repeat blob sha256:69d900227a8f4d4d2647927b9fa8da77f0f535ca497bc771c5f8a72b0cc971df
Skipping fetch of repeat blob sha256:45c9ad96a035ae2a5bda28a6e666ed7e1bbe5d945180faafd1c2ed611473e728
Skipping fetch of repeat blob sha256:f6f2416c67056dd5c3a5948858ab4578631e13cc36ee43bd8d44dea4ec4693f7
Skipping fetch of repeat blob sha256:0079bdf52f600362737f2c00e7e7ae11458844ef1d06265caf24115be990c4ab
Copying config sha256:729e7d1e2a8b59f2e885f4c7586be463050a70c00cc06bdef306e93a5bf88922
4.11 KiB / 4.11 KiB [======================================================] 0s
Writing manifest to image destination
Storing signatures
2021/01/10 11:41:34 info unpack layer: sha256:ecd924c7226f314c16de965258f37da4aa990c07e494be1116af512706138401
2021/01/10 11:41:37 info unpack layer: sha256:4fec22ce03e6be2d27efb3e9a90be68b14183859abf0864518e2581aa49fb8f5
2021/01/10 11:41:37 info unpack layer: sha256:69d900227a8f4d4d2647927b9fa8da77f0f535ca497bc771c5f8a72b0cc971df
2021/01/10 11:41:38 info unpack layer: sha256:45c9ad96a035ae2a5bda28a6e666ed7e1bbe5d945180faafd1c2ed611473e728
2021/01/10 11:41:38 info unpack layer: sha256:f6f2416c67056dd5c3a5948858ab4578631e13cc36ee43bd8d44dea4ec4693f7
2021/01/10 11:41:38 info unpack layer: sha256:0079bdf52f600362737f2c00e7e7ae11458844ef1d06265caf24115be990c4ab
INFO: Creating SIF file...
INFO: Build complete: r-base_latest.sif
The command will take a minute and create the SIF file in the directory where the apptainer command was executed (the current directory). List your directory to see the .sif file. If you pulled a specific version of R-base, your image will have a different name than that shown here.
ls -alh
474M r-base_latest.sif
You can run the R binary within the container like below. If you created the R library directory as described above (and, more generally, if you want to load or save files), don't forget to use the --bind command. You can bind anywhere deeper on the filepath than from where you need to load or save things, with /gscratch being a good option.
apptainer run --bind /gscratch r-base_latest.sif R
R version 4.4.0 (DATE) -- "Some Cute Name - Typical R Stuff"
Copyright (C) YEAR The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
> library(tidyverse)
Error in library(tidyverse) : there is no package called ‘tidyverse’
>
Note this R-base container has no packages except the R base packages. You can run install.packages() as you normally would if you were working with R locally and it will install all the files to whatever path you set R_LIBS to in the user environment instructions.
Tidyverse Container
The most popular library for R is Tidyverse (More information here), which includes packages like ggplot2, dplyr, and others. As you can see in the previous section, it doesn't exist if we use the r-base Rocker container.
Your options are to:
- run
install.packages("tidyverse")in the R-base container (r-base_latest.sif; as shown above) or - use the Rocker
tidyversecontainer with it pre-installed.
Option 1, while ok, uses a lot (and I mean a lot) of inodes as well as taking a long time to compile. It's much leaner on the cluster and faster to use a pre-built container if you know you'll use the Tidyverse.
Prior instructions on R user environment above apply. This container will also use the directory you designative in your ~/.Renviron config file. Once downloaded (the Docker to Apptainer conversion will take a few minutes), it will create a separate SIF file as shown below.
# remember to do this on a compute node
# start an interactive job with the following if you haven't yet
salloc --partition=ckpt --cpus-per-task=1 --mem=16G --time=2:00:00
apptainer pull docker://rocker/tidyverse
INFO: Converting OCI blobs to SIF format
INFO: Starting build...
Getting image source signatures
Skipping fetch of repeat blob sha256:a4a2a29f9ba48efd3d2075f395538b2eec56fb1bedfb7aecf5e54174446f9e2a
Skipping fetch of repeat blob sha256:127c9761dcbaa288abc58fc56437c2f2ffbe611b9f7f30e0b5b43cd348bb2094
Skipping fetch of repeat blob sha256:d13bf203e905463e64d89b14509aafa983fb8baf7c1931fe0a65652aeb6c838f
Skipping fetch of repeat blob sha256:4039240d2e0b4bcb42ccbce75bc54570e471ad81457478de35fbeef63536e9c0
Copying blob sha256:0ccb9f239bc673ecea30ef11ce2c495bbb85abdd96d675d44f60324c7c45d387
20.56 KiB / 20.56 KiB [====================================================] 0s
Copying blob sha256:c01d69634a9de92ab364afa9d6da377ba923131a4e170c3983a9243acafb0879
278.52 MiB / 278.52 MiB [=================================================] 11s
Copying blob sha256:d6e983dd45f0e15594336e5aabbcd004fbbb7efd8355ebd5e07f916837659fd6
225.42 MiB / 225.42 MiB [==================================================] 9s
Copying blob sha256:0d19f4c62fe952f2cec37dbde49decc2a787df40d87eecd7a8a657c04335c20b
190.84 KiB / 190.84 KiB [==================================================] 0s
Copying blob sha256:c4186f33a14ce3205196906179b6d17db22a3e9dfc9f34fedbb1f38bf2e715c9
190.39 MiB / 190.39 MiB [==================================================] 7s
Copying config sha256:22b3bdc99b5be46467e695d3639022d344d0fc4daf239a6da1f6b478ed1ee695
6.38 KiB / 6.38 KiB [======================================================] 0s
Writing manifest to image destination
Storing signatures
2021/01/10 12:28:57 info unpack layer: sha256:a4a2a29f9ba48efd3d2075f395538b2eec56fb1bedfb7aecf5e54174446f9e2a
2021/01/10 12:28:59 info unpack layer: sha256:127c9761dcbaa288abc58fc56437c2f2ffbe611b9f7f30e0b5b43cd348bb2094
2021/01/10 12:28:59 info unpack layer: sha256:d13bf203e905463e64d89b14509aafa983fb8baf7c1931fe0a65652aeb6c838f
2021/01/10 12:28:59 info unpack layer: sha256:4039240d2e0b4bcb42ccbce75bc54570e471ad81457478de35fbeef63536e9c0
2021/01/10 12:28:59 info unpack layer: sha256:0ccb9f239bc673ecea30ef11ce2c495bbb85abdd96d675d44f60324c7c45d387
2021/01/10 12:28:59 info unpack layer: sha256:c01d69634a9de92ab364afa9d6da377ba923131a4e170c3983a9243acafb0879
2021/01/10 12:29:10 info unpack layer: sha256:d6e983dd45f0e15594336e5aabbcd004fbbb7efd8355ebd5e07f916837659fd6
2021/01/10 12:29:22 info unpack layer: sha256:0d19f4c62fe952f2cec37dbde49decc2a787df40d87eecd7a8a657c04335c20b
2021/01/10 12:29:22 info unpack layer: sha256:c4186f33a14ce3205196906179b6d17db22a3e9dfc9f34fedbb1f38bf2e715c9
INFO: Creating SIF file...
INFO: Build complete: tidyverse_latest.sif
ls -alh
675M tidyverse_latest.sif
Now when you run this container's R binary you can successfully load the Tidyverse. Again, we bind above the place where our user libraries will be stored.
apptainer run --bind /gscratch tidyverse_latest.sif R
R version 4.4.0 (DATE) -- "Some Cute Name - Typical R Stuff"
Copyright (C) YEAR The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
> library(tidyverse)
── Attaching packages ─────────────────────────────────────── tidyverse 1.3.0 ──
✔ ggplot2 3.3.2 ✔ purrr 0.3.4
✔ tibble 3.0.1 ✔ dplyr 1.0.0
✔ tidyr 1.1.0 ✔ stringr 1.4.0
✔ readr 1.3.1 ✔ forcats 0.5.0
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
Warning messages:
1: replacing previous import ‘vctrs::data_frame’ by ‘tibble::data_frame’ when loading ‘dplyr’
2: package ‘purrr’ was built under R version 4.4.0
>
Success! Get on with making your pretty plots, you container superstar!
Rstudio Container and Graphical User Interface
Rstudio is an integrated development environment (IDE) for R. It's a front-end interface, historically a desktop application but it will be delivered through your browser in this instance. Rstudio will run in an Apptainer container on a compute node then be directed through the login node back to your local computer via port forwarding. In this way, you can use Rstudio on klone.
Step 1: Download Rstudio Container
First, you need the Rocker Rstudio container.
# remember to do this on a compute node
# start an interactive job with the following if you haven't yet
salloc --partition=ckpt --cpus-per-task=1 --mem=16G --time=2:00:00
# Pull the latest version of Rocker Rstudio (or the version of your choice)
# with apptainer
apptainer pull docker://rocker/rstudio
The following will prepare a .sif file called rstudio_latest.sif, but it might have another name if you pulled a different version.
Step 2: Prepare Slurm Job File
We will launch the container as a job with the command sbatch, which requests job from our job scheduler sftware called Slurm. Download our Slurm job file from this hyperlink which was adopted for klone from the tutorial by Rocker More information about the original tutorial can be found here.. The command below will download the file to your current directory.
wget https://hyak.uw.edu/files/rstudio-server.job
Remember to replace the word UWNetID in the paths below with YOUR username/UWNetID.
You will need to modify a few environment variables in rstudio-server.job related to R. Use nano or vim to edit the contents of rstudio-server.job:
- The
RSTUDIO_CWDpath, is your working directory, as if you were using the functionsetwd()withinR.rstudio-server.jobshows this as/gscratch/scrubbed/UWNetIDYou must change this line for this to work. We recommend setting this to the directory where you are storing your data for your intended project. Additionally, it might simplify matters if this is the folder where the container is located and downloaded to using theapptainer pullcommand above. - Set your
RSTUDIO_SIFvariable, this is name of the container file. In this case,rstudio_latest.sif. - (Optional) Set your
R_LIBS_USERpath, which inrstudio-server.jobisR_LIBS_USER=${RSTUDIO_CWD}/Ror/gscratch/scrubbed/UWNetID/RbecauseRSTUDIO_CWD="/gscratch/scrubbed/UWNetID", remember? Change these variables to fit your needs. That means for this Rstudio session my package libraries (when I useinstall.packages()) will be stored in/gscratch/scrubbed/UWNetID/R. In this case, I am matching this Rstudio session to my preferences set above in the user environment section. For your session, you might decide to designate a different directory for your R package libraries. Rememeber directories don't exist until you make them.
Additionally, you might decide to modify the sbatch directives to adjust the resources to request for your Slurm Rstudio job. For example, fill in your specific partition if applicable (check your options with hyakalloc). Also set your job run limits, cores (i.e., ntasks), memory, etc.
Review the highlighted sections of rstudio-server.job below and edit your version to fit your needs and paths you have access to:
#!/bin/sh
#SBATCH --job-name=rstudio-server
#SBATCH --partition=ckpt #update this line - use hyakalloc to find partitions you can use
#SBATCH --time=02:00:00
#SBATCH --nodes=1
#SBATCH --ntasks=4
#SBATCH --mem=20G
#SBATCH --signal=USR2
#SBATCH --output=%x_%j.out
# This script will request a single CPU with four threads with 20GB of RAM for 2 hours.
# You can adjust --time, --nodes, --ntasks, and --mem above to adjust these settings for your session.
# --output=%x_%j.out creates a output file called rstudio-server_XXXXXXXX.out
# where the %x is short hand for --job-name above and the X's are an 8-digit
# jobID assigned by Slurm when our job is submitted.
RSTUDIO_CWD="/gscratch/scrubbed/UWNetID" # UPDATE THIS LINE
RSTUDIO_SIF="rstudio_latest.sif" # update this line
###
### Truncated to save space on the web.
###
export R_LIBS_USER=${RSTUDIO_CWD}/R
Step 3: Start the Rstudio Server
Next's we'll submit the job with sbatch which will launch the Rstudio container, and then we will use port forwading to interact with the RStudio interface on our web browser.
sbatch rstudio-server.job
Submitted batch job 12345678
# Slurm will assign a JobID when the job was submmitted
# it will likely be an 8-digit number, but not 12345678
Monitor the job with squeue and your UWNetID like the following example:
squeue -u UWNetID
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
12345678 ckpt rstudio UWNetID R 3:15 1 n3088
Slurm will save your output file called rstudio-server_12345678.out in the directory where the sbatch command was executed. The suffix matches the job number you see. Check out its contents like below for instructions on how to connect to your Rstudio session.
cat rstudio-server_12345678.out
1. SSH tunnel from your workstation using the following command:
ssh -N -L 8787:n3164:47101 UWNetID@klone.hyak.uw.edu
and point your web browser to http://localhost:8787
2. log in to RStudio Server using the following credentials:
user: UWNetID
password: 410lzxMwV9EObv7aDEjm
When done using RStudio Server, terminate the job by:
1. Exit the RStudio Session ("power" button in the top right corner of the RStudio window)
2. Issue the following command on the login node:
scancel -f 12345678
The credentials are randomly generated for each sbatch job adding additional cybersecurity with a new session password each time you launch Rstudio this way.
Step 4: Start Port Forwarding
This next section is done on your local computer not on the cluster.
In a new terminal or command prompt on your laptop copy and paste the other SSH command from the Slurm output. The following is an example:
ssh -N -L 8787:n3164:47101 UWNetID@klone.hyak.uw.edu
... provide UWNetID password
... Duo 2 Factor Authentication
The login will appear to hang, but your connection is now open. If you are disconnected and reconnect you can resume your Rstudio session.
Do not use the rstudio-server password to open the ssh tunnel. After your ssh command, your UWNetID password is required. Multiple failed login attempts will result in a IP ban.
Next, open a new browser window to http://localhost:8787 and provide the password from the output file (rstudio-server_12345678.out and 410lzxMwV9EObv7aDEjm in this example).
Once you log in you should see an environment similar to the below. Both your Home directory and gscratch folders will be mounted.
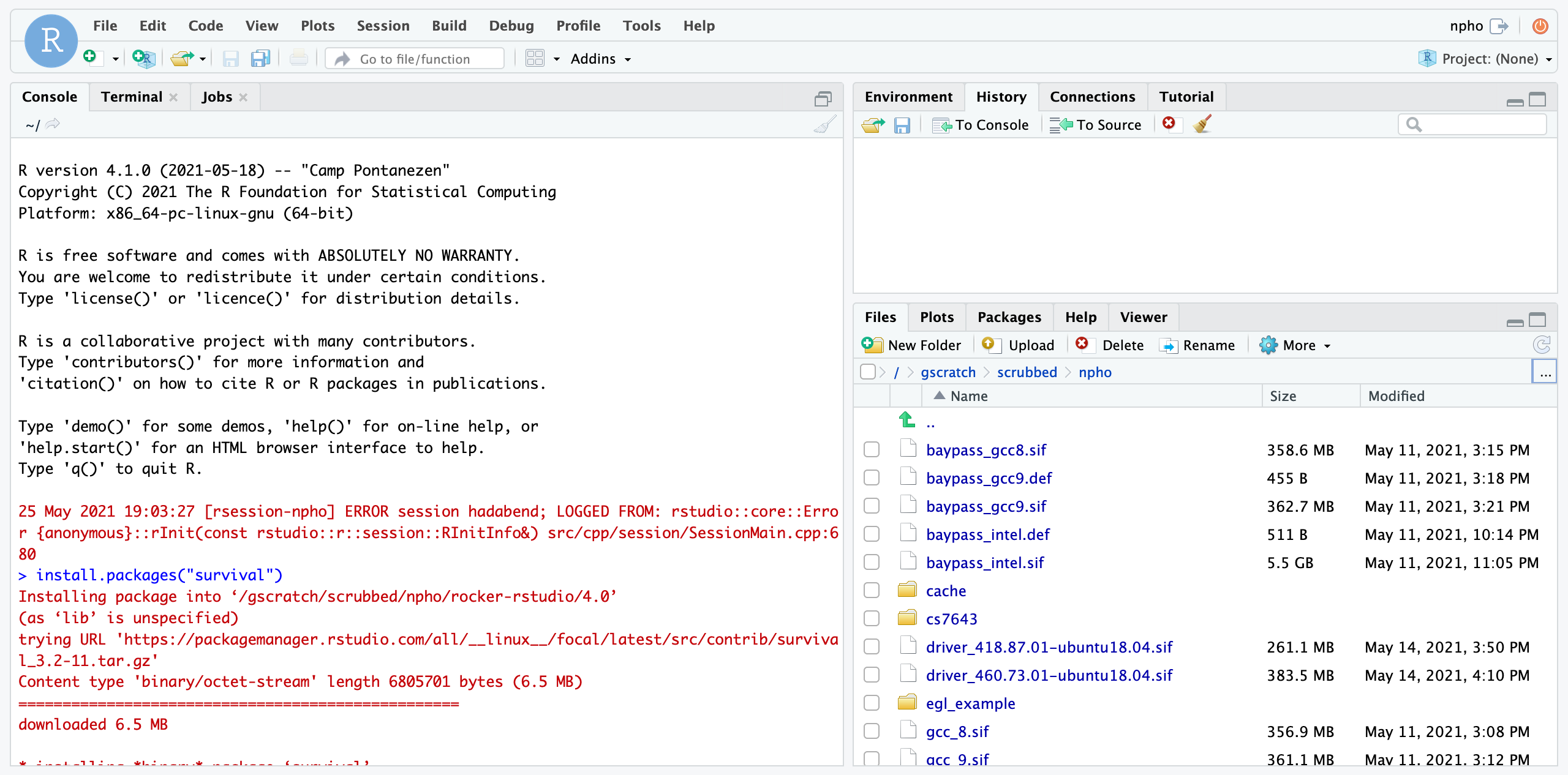
Step 5: End your Session
If you did not adjust the --time directive in rstudio-server.job, your session will end after 2 hours.
Preferably, you can end your session manually. Exit the RStudio Session ("power" button in the top right corner of the RStudio window). Then go back to klone and use the scancel command provided with the specific jobID. For example,
scancel -f 12345678
Regular use of this method
Once you are satisfied with the job settings and configuration of your Rstudio session, you can reuse this method everytime you want to use Rstudio by starting at Step 3: Start the Rstudio Server above.
If you have trouble with this method, please report errors in an email to help@uw.edu with Hyak in the message.
R via Modules
There are some versions of R still available as modules. Use these at your own risk. They may be versions with deprecated packages, and many were contributed by other users who built them to fit their personal needs, not yours. The Hyak team will not provide support for the use of these modules.
module avail
----- /sw/modules-1.775/modulefiles -----
r_3.3.3 r_3.5.1 r_3.6.0 r_3.6.0+Rmpi-impi_2019
----- /sw/modules-1.775/modulefiles -----
contrib/r/3.4.3 contrib/r/3.5.1 contrib/r/3.6.1