Rstudio
R is a language used for statistical computing and data analysis. For ease of use, R can be used via Rstudio, an integrated development environment (IDE) that supports both R and Python languages. For more information, refer to the documentation pages for R and Rstudio. The following section will provide a walkthrough for using R and Rstudio on Hyak via Open Ondemand (OOD).
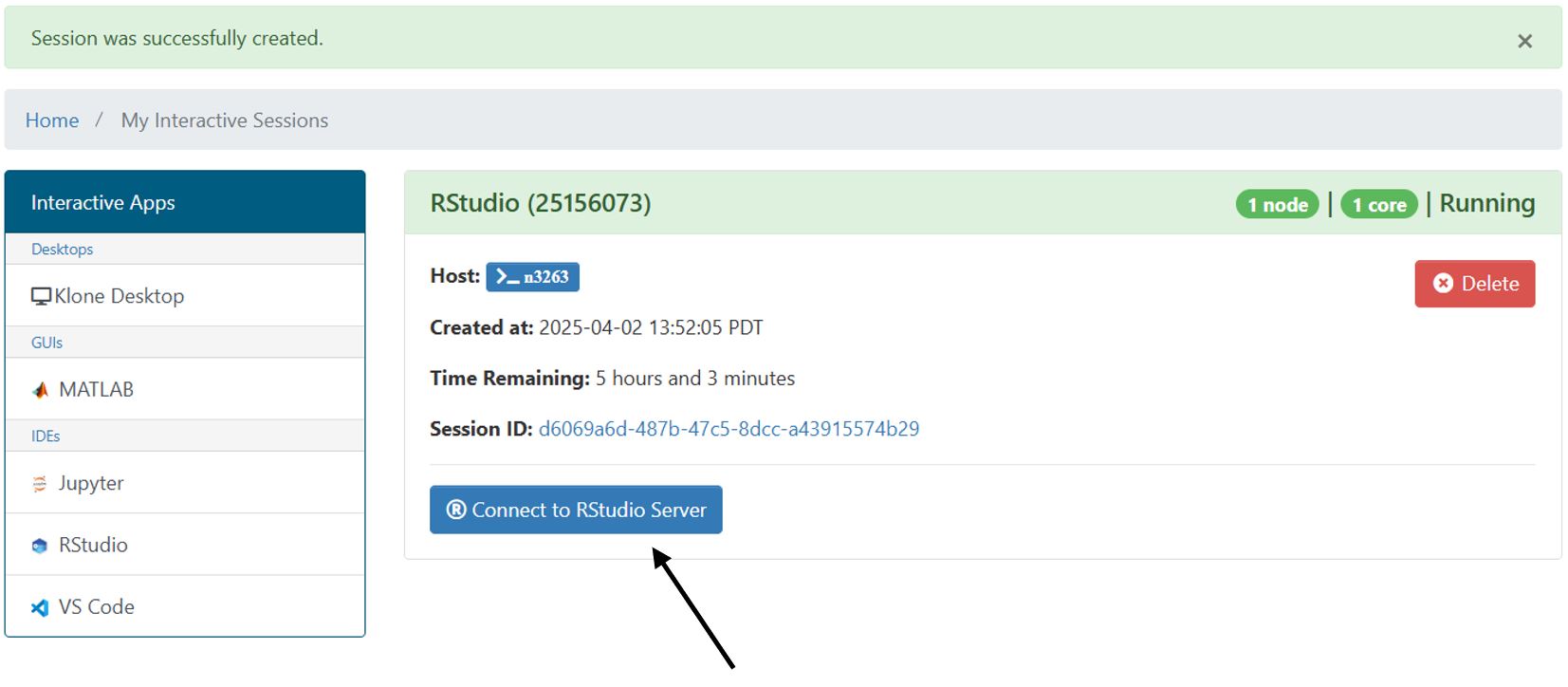
Rstudio Interactive App
To access OOD off campus, you must connect to a VPN. For more information on working remotely, click HERE.
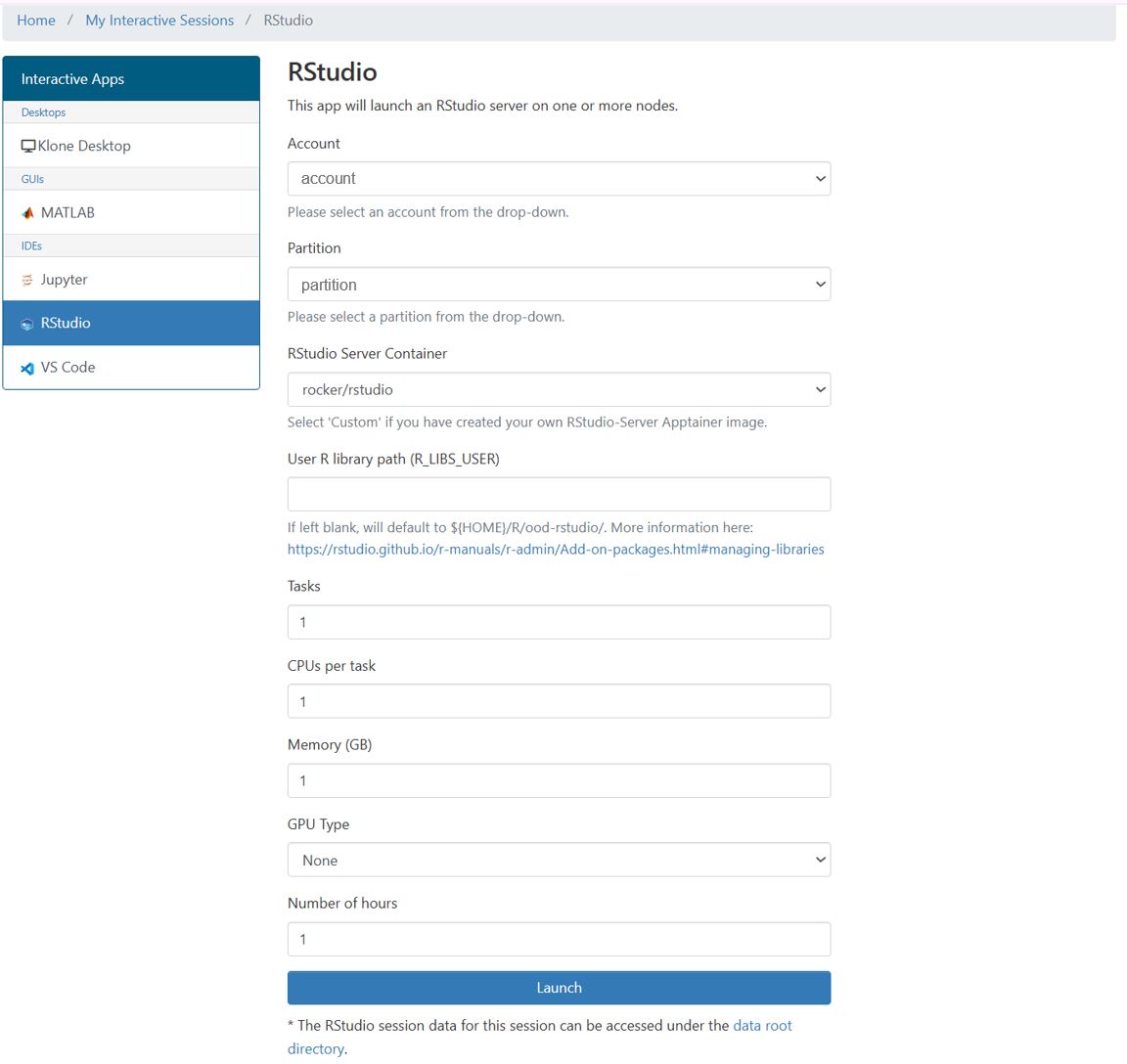
To access the Rstudio interactive app, navigate to the OOD portal HERE and select Rstudio from the drop down menu of interactive apps from the dashboard at the top of the page. Similar to Hyak's job scheduler Slurm, this form allows you to select the account and partition for your job. To use community idle resources, select the ckpt partition.
You are also able to select a Rstudio server container that is provided from the rocker project. Alternatively, you may use a custom container by selecting "Custom" and specifying the absolute path to the container (e.g. /mmfs1/sw/ondemand/containers/rstudio/sifs/tidyverse-27jan2025.sif). Below we demonstrate how to build your own custom R container.
Once you specify the Rstudio server container, you have the option to change the User R Library Path (R_LIBS_USER). When you use the install.packages() command, the package library along with all of its dependencies will be installed in the directory specified with the User R Library path (R_LIBS_USER) field. This path defines the library location where R stores packages. By default, this is the path to your home directory which has a 10G storage limit. If you plan on using install.packages(), it is recommended to change the User R Library Path to somewhere with a larger storage quota to install libraries.
Custom R Containers
The install.packages() command can be used to install most packages during your Rstudio interactive session as long as the container has all the dependencies required by the package. If your container environment is properly set up, install.packages() will run without issues. Some packages may require changes to the operating system. To install these packages, you will need to build a custom container. Because R is open source and the dependencies are constantly changing, using custom R containers will help maintain the R version and all dependencies in an isolated environment. This way, you can maintain access to tools you rely on if packages and versions become unsupported. Because R can take up a large amount of file storage, containers are also useful for reducing your storage usage. The following section will provide a walkthrough of building a custom container to run R in.
To start off, create a definition file to build your container. This may look something like the following:
Bootstrap: docker
From: rocker/tidyverse
%post
# Update Ubuntu packages
apt-get update -y
# Installing Ubuntu system libraries
# Listed below are common examples of Ubuntu OS libraries that may be missing
apt-get install -y libglpk40
apt-get install -y \
libxml2 \
libxt6 \
zlib1g-dev \
libbz2-dev \
liblzma-dev \
libpcre3-dev \
libicu-dev \
libjpeg-dev \
libpng-dev \
libxml2-dev \
libglpk-dev \
libz-dev
# Installing CRAN packages via Rscript into the container
Rscript -e 'install.packages("RColorBrewer")'
# Installing Bioconductor packages
# The BiocManager package is required to install Bioconductor packages
if (!require("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install(version = "3.20")
# You can now install any Bioconductor packages using BiocManager
Rscript -e 'BiocManager::install("regioneR")'
Rscript -e 'BiocManager::install("regioneReloaded")'
# Recommended for use with OOD - setting up symbolic links that match klone's filesystem
mkdir /scr /mmfs1
ln --symbolic /mmfs1/sw /sw
ln --symbolic /mmfs1/data /data
ln --symbolic /mmfs1/gscratch /gscratch
The above example serves as a template and should be edited to fit your R environment needs.
Once your definition file is created, you can build the container:
apptainer build customR.sif customR.def
In this example, the latest version of rocker/tidyverse is being pulled.
If you would like to use a specific version of R, you can
change From: rocker/tidyverse at the start of your .def file to something like From: rocker/tidyverse:4.3.2.
Because the tidyverse package includes a large collection of R packages that
would be time consuming to install from scratch,
using a prebuilt rocker/tidyverse image is generally a good choice for a
base container.
Some prebuilt containers are tailored for specific types of workflows and can save time setting up complex environments:
-
Rstudio server container from Tufts University HPC. This container has various CRAN and Bioconductor packages installed.
-
Single Cell RNA-seq container from Tufts University HPC. This container has a large range of bioinformatics packages that are useful for scRNseq analysis.
-
Rocker project containers. These containers are useful for running R and Rstudio Server environments with a variety of preinstalled packages. For more information regarding the specifications of each container, please refer to the Rocker Project website.