August 2025 Maintenance Update
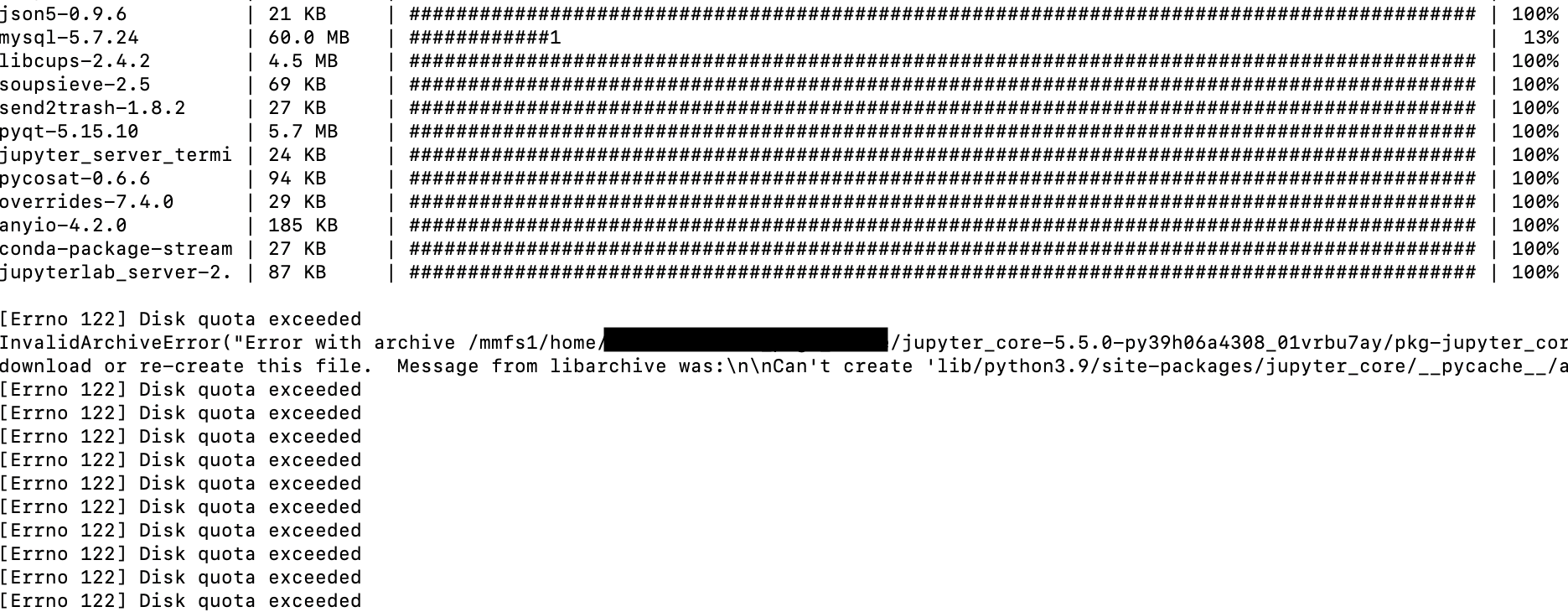
During August’s maintenance, we refreshed the operating system images for both login and compute nodes, upgraded Slurm to version 25.5.2, and upgraded Klone's filesystem (GPFS) for increased stability. We also introduced a new Globus OneDrive connector, making it easier than ever to transfer files between OneDrive and Hyak Klone or Kopah Storage.
Stay informed by subscribing to our mailing list and the UW-IT Research Computing Events Calendar. The next maintenance is scheduled for Tuesday, September 9, 2025 (the second Tuesday of the month).
Notable Updates
- Node image updates – Routine updates plus installation of new Slurm utilities that we will test for job efficiency monitoring.
- Slurm upgrade to 25.5.2 – Resolves a bugs the
--gresflag allowed resources to be allocated to more than one job in version 25.05.0 and fixes X11 forwarding. Read more about this version. - GPFS upgrade to 5.1.9.11 – Improves stability and includes several bug fixes. Read more about this version.
New Features
Globus OneDrive Connector – UW-IT Research Computing has added OneDrive as a connector to Globus, making transfers between OneDrive and Hyak Klone or OneDrive and Kopah Storage easier than ever before!
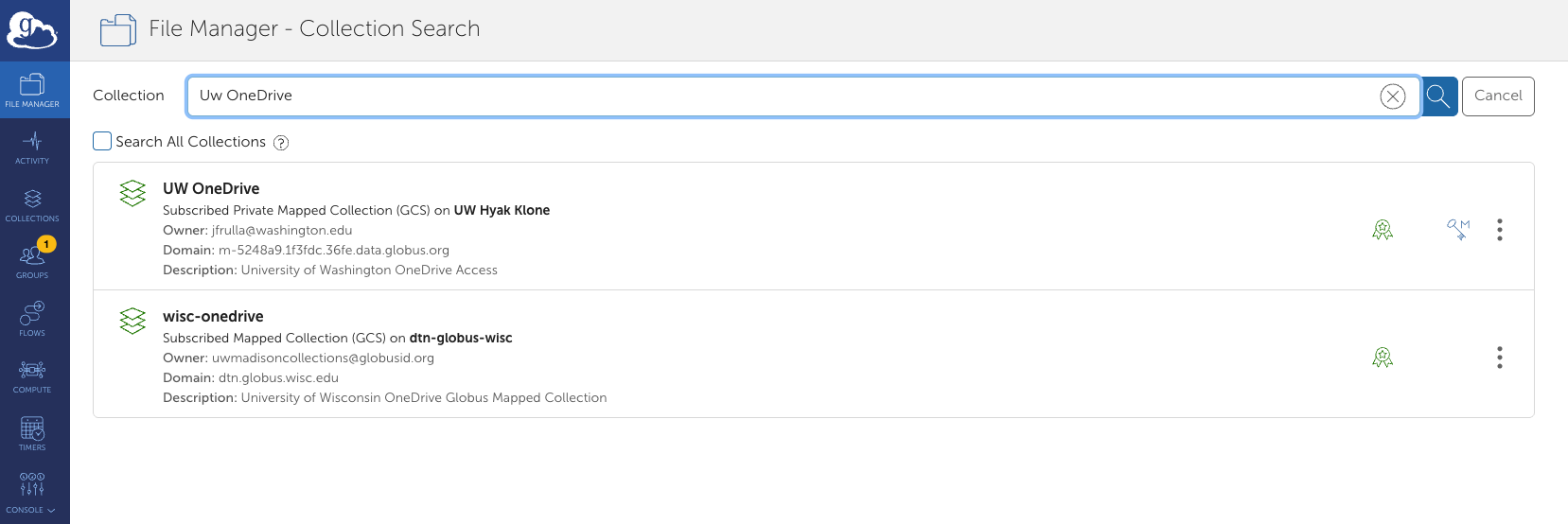
Things to note
- Did you know that the UW Community are eligible for 5TB of storage on OneDrive as part of the Office 365 Suite? Click here to learn more.
- While OneDrive is HIPAA and FERPA compatible, encryption is not enforced for Globus transfers on any of our connectors (OneDrive, Kopah, Klone). As a reminder, Klone and Kopah are NOT aligned with HIPPA, please keep this in mind now that OneDrive can transfer to either cluster.
- Sharing with external partners is not enabled for our OneDrive or Klone connectors via Globus. Sharing is permitted for Kopah.
- Read more
Office Hours
- Wednesdays at 2pm on Zoom. Attendees need only register once and can attend any of the occurrences with the Zoom link that will arrive via email. Click here to Register for Zoom Office Hours.
- Thursdays at 2pm in person in eScience. (address: WRF Data Science Studio, UW Physics/Astronomy Tower, 6th Floor, 3910 15th Ave NE, Seattle, WA 98195).
- See our office hours schedule, subscribe to event updates, and bookmark our UW-IT Research Computing Events Calendar.
If you would like to request 1 on 1 help, please send an email to help@uw.edu with "Hyak Office Hour" in the subject line to coordinate a meeting.
UW Community Opportunities
- The Data Science and AI Accelerator pairs eScience Institute data scientists with researchers from any field of study to work on focused, collaborative projects. Collaborations may center on analysis of an existing dataset to answer a specific research question, an implementation of software for processing or analyzing data, data visualization tools, or tools for data interpretation. Accelerator Projects may be submitted at any time. Projects for Fall 2025 must be received by August 14th, 2025. LEARN MORE HERE.
- Applications for the CSDE Data Science and Demography Training program are due Friday, August 22nd by 5pm. An information session will take place Wednesday, August 13th at 10:00 a.m. DETAILS HERE.
- Cloud Clinic August 14 10-11am - guest presenter Niris Okram from AWS presenting on “The Utility of Capacity Blocks: Optimizing computing horsepower per budget dollar.” This will be followed by a short presentation on building small-scale (“Littlest”) JupyterHubs. LEARN MORE HERE.
- DubHacks - October 18 - October 19, 2025 - DubHacks 2025 takes you back to where it all began—the childhood bedroom. A space for imagination, curiosity, and bold ideas. Now, with code instead of crayons, you get to build what makes your younger self proud. No limits, just pure creativity. What will you create when you let yourself play?
External Training Opportunities
- Automating Research with Globus: The Modern Research IT Platform - Aug. 18, 2025, 9 a.m. – 12 p.m. (Pacific Time) This workshop introduces Globus Flows and its role in automating research workflows. Participants will explore data portals, science gateways, and commons, enabling seamless data discovery and access. Enroll here.
- CU-RMACC Webinars: Should I be Scared of AI? Aug. 18, 2025 - 3:00 PM - 4:00 PM EDT Throughout history, new technologies have sparked both excitement and fear—AI is no different. In this talk, Dr. Shelley Knuth, Assistant Vice Chancellor for Research Computing at the University of Colorado explores the common fears surrounding artificial intelligence, why we feel them, and how we can shift our perspective to focus on positive outcomes. We’ll look at practical ways to address risks, embrace innovation, and move forward with AI as a powerful tool rather than something to fear. Learn more and register.
- COMPLECS: Batch Computing (Part II): Getting Started with Batch Job Scheduling 08/21/25 - 2:00 PM - 3:30 PM EDT Learn more and register.
- NUG Community Call: A Birds-Eye View of Using Cuda with C/C++ on Perlmutter (Part 2) August 27, 2025, 11 a.m. - 12:30 p.m. PDT - In this two-part training series, users will be introduced to the basics of using CUDA on Perlmutter at NERSC. The training will focus on the basics of the Perlmutter architecture and NVIDIA GPUs, programming concepts with CUDA using C/C++. Event 2 focuses on advance kernel and custom cuda kernels in C/C++. Learn more and register.
- COMPLECS: Linux Tools for Text Processing 09/04/25 - 2:00 PM - 3:30 PM EDT Learn more and register.
- Python for HPC 09/09/25 - 2:00 PM - 3:30 PM EDT Learn more and register.
- COMPLECS: Data Transfer 09/18/25 - 2:00 PM - 3:30 PM EDT Learn more and register.
- COMPLECS: Interactive Computing 10/09/25 - 2:00 PM - 3:30 PM EDT Learn more and register.
- COMPLECS: Linux Shell Scripting 10/23/25 - 2:00 PM - 3:30 PM EDT Learn more and register.
- COMPLECS: Using Regular Expressions with Linux Tools 11/06/25 - 2:00 PM - 3:30 PM EST Learn more and register.
- COMPLECS: Batch Computing (Part III) High-Throughput and Many-Task Computing - Slurm Edition 12/04/25 - 2:00 PM - 3:30 PM EST Learn more and register.
- R for HPC 12/04/25 - 2:00 PM - 3:30 PM EST Learn more and register.
Positions
- Two PhD positions in Artificial Intelligence - in collaboration with German Aerospace Center and TU Dresden, Germany. Deadline to apply: 27 August 2025. Apply Now!
Questions about Hyak Klone, Tillicum, or any other UW-IT Research Computing Service? Fill out our Research Computing Consulting intake form. We are here to help!
Happy Computing,
Hyak Team